Biology Research Groups
Research Groups
B J Rao
My programme relocated to IISER-Tirupati from TIFR-Mumbai very recently. I continue my research efforts at IISER-Tirupati that are mechanism-centric and are mainly directed towards two questions of cellular-molecular biology.
- How do chromosomes maintain quasi-stability against a variety of cellular stresses? We probe signaling mechanisms that lead to such genome-homeostasis in mammalian cells.
- Chlamydomonas reinhardtii, single celled plant organism exhibits tight coordination between chloroplast and mitochondrial compartments of a cell. How is this tuned under changing cellular states? We hope to glean the dynamic signaling cross-talk between these organelles which is unique to plant world and hope to extract new Biology from it.
Molecular Microbiology Laboratory (Raju Mukherjee)
The focus of our research is circumscribed around understanding the biology of Mycobacterium species with an ultimate objective of developing strategies for successful interventions to stop the diseases (e.g. Tuberculosis) caused by these microorganism.
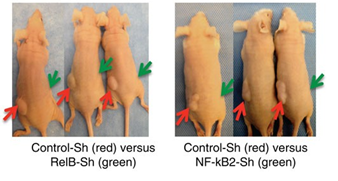
Sivakumar Vallabhapurapu Reseach Area:
We work in the areas of Cancer Biology and Immunology. The focus is on how different signaling pathways regulate specific gene expression signatures in different cancer cell types and in the immune cells. While our major focus is on NF-kB signaling, which is abnormally activated in many cancer types and is the Central Pillar of the Immune system, we also work on different other signaling pathways such as the MAPK, AKT, mTOR etc. Further, we try to understand how the cross-talk between such signaling pathways contribute to cancer progression and at the same time also contribute to normal homeostatic regulation in the Immune System. Role of different signaling pathways in normal cancer cells and cancer stem cells will be studied. We employ transgenic, knockout and other mouse models as well as invitro Molecular Biology, Cell Biology and Biochemical expertiments (such as protein-protein interactions) for our study. For a respresentative figure of our study on NF-kB signaling in the progression of Multiple Myeloma please see the Figure.
Plant Development & Applied Genetics Group (Eswar Rami Reddy)
Our group focuses on deciphering the molecular mechanisms and the regulatory systems that enable plants to orchestrate complex root system architecture. Further, improvement of tolerance in crop plants to abiotic stresses (drought, salt and heavy metal) by improving root traits.
Molecular Oncology and Cell Signalling (Pakala Suresh Babu)
Our group works on understanding the molecular mechanisms of coregulators in signal transduction, Gene regulation, inflammation, Cancer progression and Cancer metastasis. Our research interests also include developing transgenic and knock out mouse models for studying the role of coregulator signalling in cancer and inflammatory disorders.
Cellular and Molecular Parasitology (Suchi Goel)
Our research focuses on pathogenesis of severe malaria parasite where adhesion of parasites in the microvasculature leads to activation of stress signalling pathways in human host.
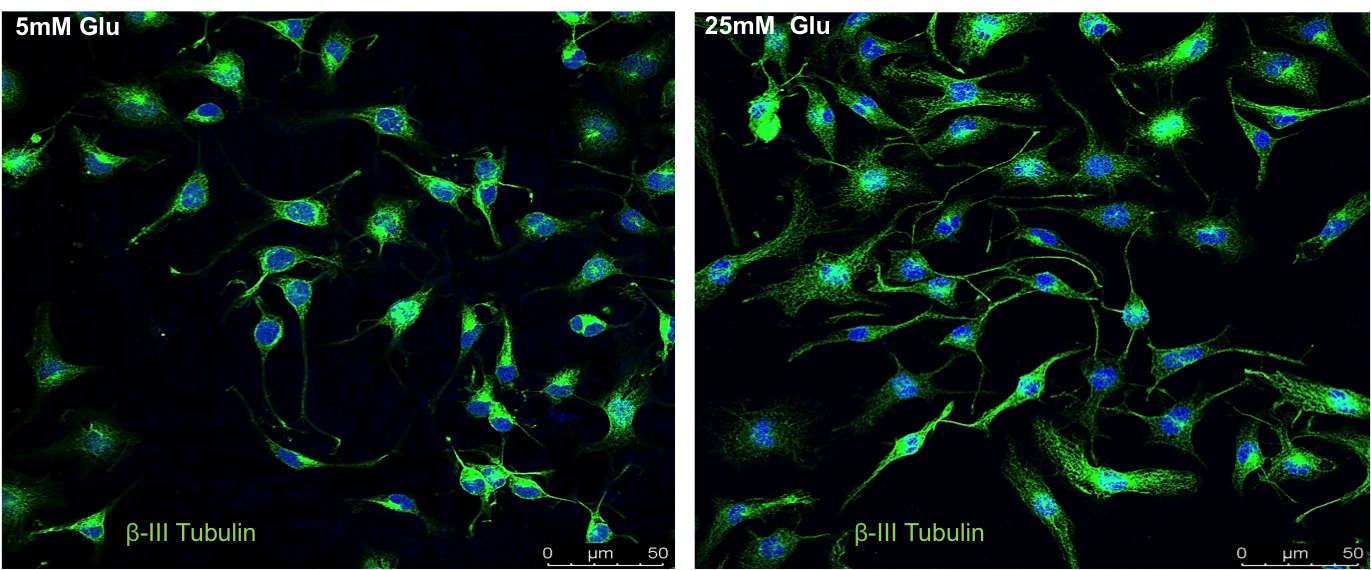
Neuronal Metabolism lab (Vasudharani Devanathan)
Cellular and Vascular Biology Lab (Sanjay Kumar)
Our lab research focuses on signaling mechanisms that define the complex responses to TGF-β in the mammalian system. We also study the role of mitochondrial proteins in normal cellular functions and human diseases.
Ash Lab (Ashwani Sharma)
Ash Lab works in the area of nucleic acid aptamers, which are single stranded DNA or RNA oligonucleotides having high affinity and specificity to their target. Aptamer binding to its target is utilized in designing new biosensing platforms for disease detection and diagnostics. In addition, the lab also focuses on construction of DNA/RNA based nanoparticles decorated with aptamers and oligotherapeutics such as siRNA for the targeted drug delivery to different cancers.

Plant Stress Biology group (Annapurna Devi Allu)
Our research focuses on understanding the transcriptional and epigenetic regulation of plant growth and stress responses. We are particularly interested in studying the effect of priming on plant response to individual and combined environmental stresses. The molecular mechanisms that determine the duration of primed state and stress memory in plants will be investigated.
Single Molecule Biophysics Lab (Nibedita)
Our research focuses on amalgamating DNA nanotechnology and biomolecule in its native form to answer basic questions that are pertinent to the field of Biophysics. In particular we are interested in designing reconfigurable DNA structures responsive to external stimuli and its potential application in therapeutics. We are also interested in exploring the mechanism of biomolecule induced conformational transitions of higher order DNA junctions and structures using single molecule techniques such as Total Internal Reflection Fluorescence Microscopy, Fluorescence Correlation Spectroscopy, multi-parameter photon time-stamping spectroscopy etc.
Genomics and Systems Biology group (Sreenivas Chavali)
Our research aims at obtaining a holistic understanding of how variation in information flow affects phenotypes and fitness. We particularly focus on understanding how sequence variation- single nucleotide changes, repeats or compositional bias within intrinsically disordered proteins/regions influence the function(s), regulation and evolution of biomolecules, with special emphasis on health and disease. To address this, we investigate biological information flow at different scales of complexity (genome(s), systems and molecular levels) and at different time-scales (at an instant to millions of years) in diverse organisms ranging from yeast to humans. Our research work involves computational investigations integrating and analyzing large-scale datasets, coupled with experimental investigations, primarily using budding yeast as a model system.
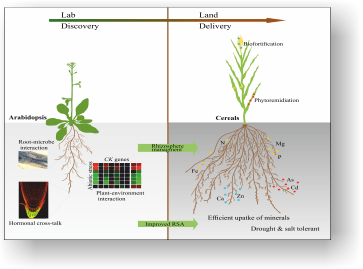
Plant Signaling Group (Swarup Roy Choudhury)
We are interested in identifying the molecular and cellular mechanisms behind signal transduction in plants to unravel plant signaling pathways extending from receptors to signaling cascades and target genes that are crucial to root nodule development, plant architecture determination and stress adaptation. The important questions are: How signaling is initiated by receptor-like kinases and which kind of ligands bind to the receptors? How is downstream regulatory network modulated by receptor kinases from lower to higher groups of plants? How is receptor-mediated signaling orchestrated the gene expression? We approach our questions both in model and crop plants with current tools in biochemistry, molecular biology, genetics, cell biology and biotechnology to achieve a notion for improving agricultural productivity.
Chromosome Integrity and Inheritance Lab (Viji Subramanian)
Our research is focused on addressing two outstanding questions in the field.
1.How are DNA breaks distributed to short chromosomes at a higher density despite their short size to promote inheritance fidelity?
2.What molecular mechanisms govern the repair template choice between sister chromatid and the homologue to ultimately promote genome integrity and inheritance?