The Bar research group mainly focuses on the cutting-edge research to address the problems, ranging from the very fundamental to applications, of molecular and supramolecular magnetic materials. Multi-pronged chemical strategies are employed to improve the magnetic properties in view of the applications in modern technologies. The research group designs and synthesizes smart organic/inorganic ligands judiciously… Continue reading Single-Molecular Nanomagnets (Arun Kumar Bar)
Category: Chemistry
Ash Lab (Ashwani Sharma)
Ash Lab works in the area of nucleic acid aptamers, which are single stranded DNA or RNA oligonucleotides having high affinity and specificity to their target. Aptamer binding to its target is utilized in designing new biosensing platforms for disease detection and diagnostics. In addition, the lab also focuses on construction of DNA/RNA based nanoparticles… Continue reading Ash Lab (Ashwani Sharma)
Energy Storage Materials (Vanchiappan Aravindan)
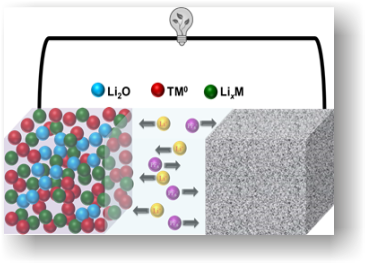
We perform research activity in the Materials Chemistry, specifically energy storage materials for batteries and supercapacitors. Our prime aspect of work is to realize the high energy and power capability in a single device by integrating rechargeable batteries and supercapacitors. Recycling spent batteries and bio-waste derived material towards charge storage devices to keep our environment… Continue reading Energy Storage Materials (Vanchiappan Aravindan)
Main Group Chemistry & Catalytics (Sudipta Roy)
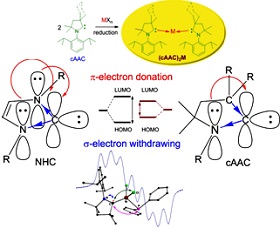
Our group is interested in the syntheses of carbene stabilized zero-valent heterodiatomic compounds with main group elements and/or mixed transition metal/main group elements and studying their physical and catalytic properties for the syntheses of biologically relevant small organic molecules
Protein Nanomechanics lab (Soumit Sankar Mandal)
The main focus of our research group is to investigate the correlation between the structure and function of multidomain proteins using Spectroscopic and Single Molecule Optical tweezers technique.
Banerjee Research Group (Shibdas Banerjee)
We work on clinical mass spectrometry imaging for cancer diagnosis and disease biomarker discovery. Our laboratory also focuses on methodology developments using ambient ionization mass spectrometry to explore natural product discovery, microdroplet chemistry, catalysis and mechanistic organic chemistry.
Computational and Quantum Chemistry (Raghunath O Ramabhadran)
Our group primarily works in the area of computational and quantum chemistry. We eagerly work towards new developments in application of computational tools/protocols in organic/bio-organic, inorganic, and physical chemistry. As an auxiliary area, we are also interested in scientific pedagogy.
Biomimetic Systems and Bioinorganic Chemistry group (Pankaj Kumar Koli)
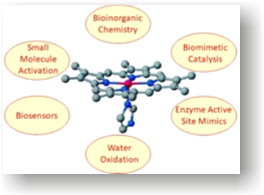
Our group mainly focus on biomimetic systems and small molecule activation (O2, NO, CO, CO, and H2S, etc.) in the field of bioinorganic chemistry. In addition, we also focus on bio-sensing, catalysis and insight into the mechanistic pathways of various enzymatic reactions.
Catalysis and Synthesis Group (Gopinath Purushothaman)
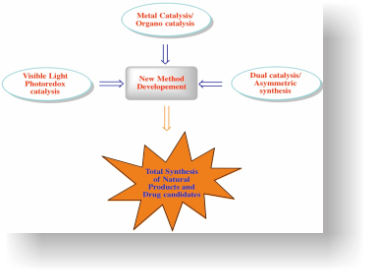
Our primary research focus is to develop new synthetic methods using catalysis as the main tool with special focus on visible light photoredox catalysis, transition metal catalysis & asymmetric catalysis. These synthetic methods will later be used for applications in total synthesis of natural products, drugs & other bioactive compounds.
Electrochemistry of Materials (Prof. Vijayamohanan Pillay)
I work on a variety of problems at the interface between Electrochemistry and Materials Chemistry, sometimes using electric filed to make unusual, two-dimensional materials like graphene and phosphorene quantum dots while in some other cases to make hybrid materials for applications ranging from more efficient electrocatalysts for energy storage (e.g.,fuel cells and batteries) to self-assembled… Continue reading Electrochemistry of Materials (Prof. Vijayamohanan Pillay)